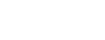
University of Texas at Austin
Dept, of Chemistry & Biochemistry
105 E. 24th St. Stop, A5300
Austin, TX 78712-0165
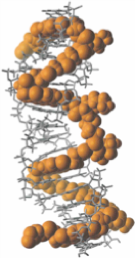
After studying the three-dimensional structure of a protein or DNA, one cannot help but marvel at the large size, complexity and asymmetry of biological molecules; all of which enable a remarkable array of functions such as precise recognition (i.e. antibodies), exquisite catalysis (i.e. enzymes) and information storage (i.e. DNA) to name a few. These remarkable properties are only possible because non-covalent interactions control folding and assembly of biological molecules sufficient to overcome conformational chaos.
Historically, the field of organic chemistry has focused on controlling molecular structure by creating new covalent bonds. Organic chemistry is evolving in many new and exciting directions, and among them are attempts to control molecular structure through the harnessing of non-covalent forces. Once harnessed, non-covalent interactions will hold the key to developing engineered molecular systems capable of important new functions that are far beyond the capacities of even the most sophisticated synthetic molecules of today. Research in the Iverson group is centered on the construction and manipulation of large molecular systems in water, ranging from abiotic molecules to proteins.
CHEMISTRY
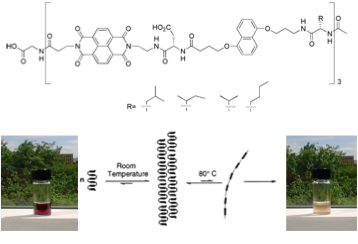
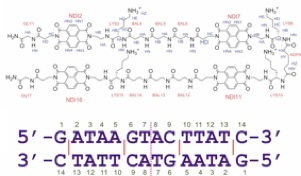
Students or post-doctoral fellows working on these projects will acquire a comprehensive knowledge of computer guided molecular design, organic synthesis, and especially the manipulation of non-covalent interactions in water or the solid state. Depending on the project, they will also become experts in nucleic acid chemistry, materials science or photovoltaic technology. Recent graduates from the laboratory have been successful in academic positions as well as industry because the focus on the lab is on innovation and the control of molecular systems in aqueous solution, both of which are of growing interest to academic researchers and companies alike.
MOLECULAR BIOLOGY

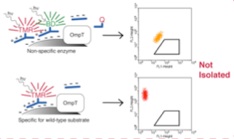


vs.
Control
The Chemistry of Large Molecules in Water
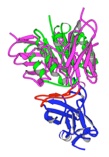
M18 Antibody-PA
Co-crystal
OmpT: E. coli surface protease that strongly prefers Arg-arg cleavage sites